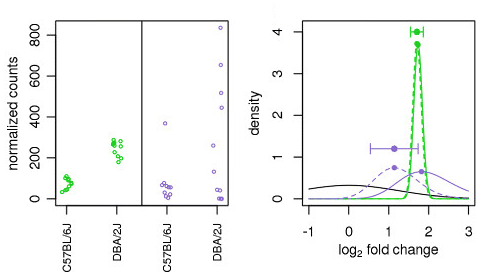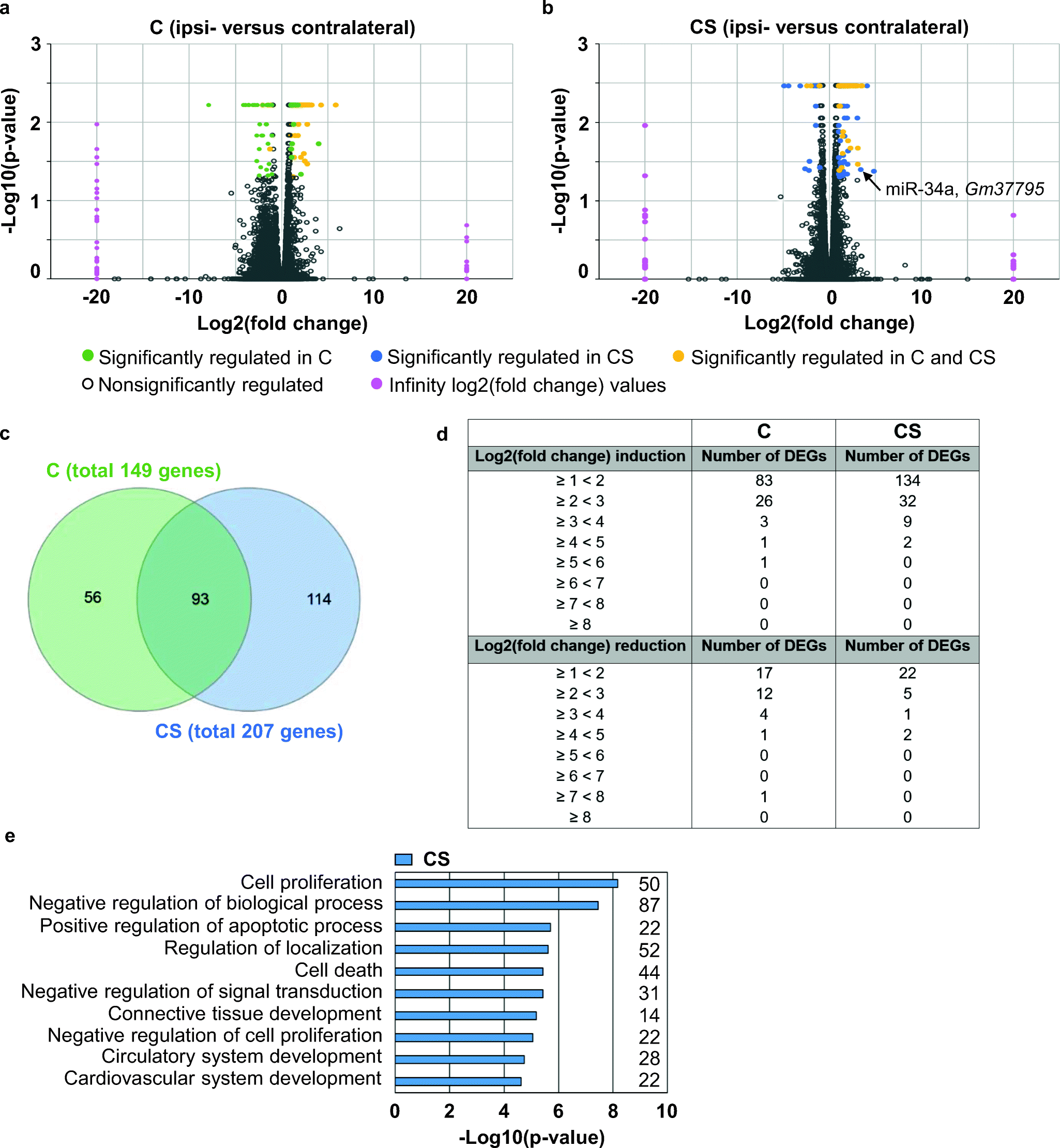
Figure 3 | Endothelial Cell-Specific Transcriptome Reveals Signature of Chronic Stress Related to Worse Outcome After Mild Transient Brain Ischemia in Mice | SpringerLink

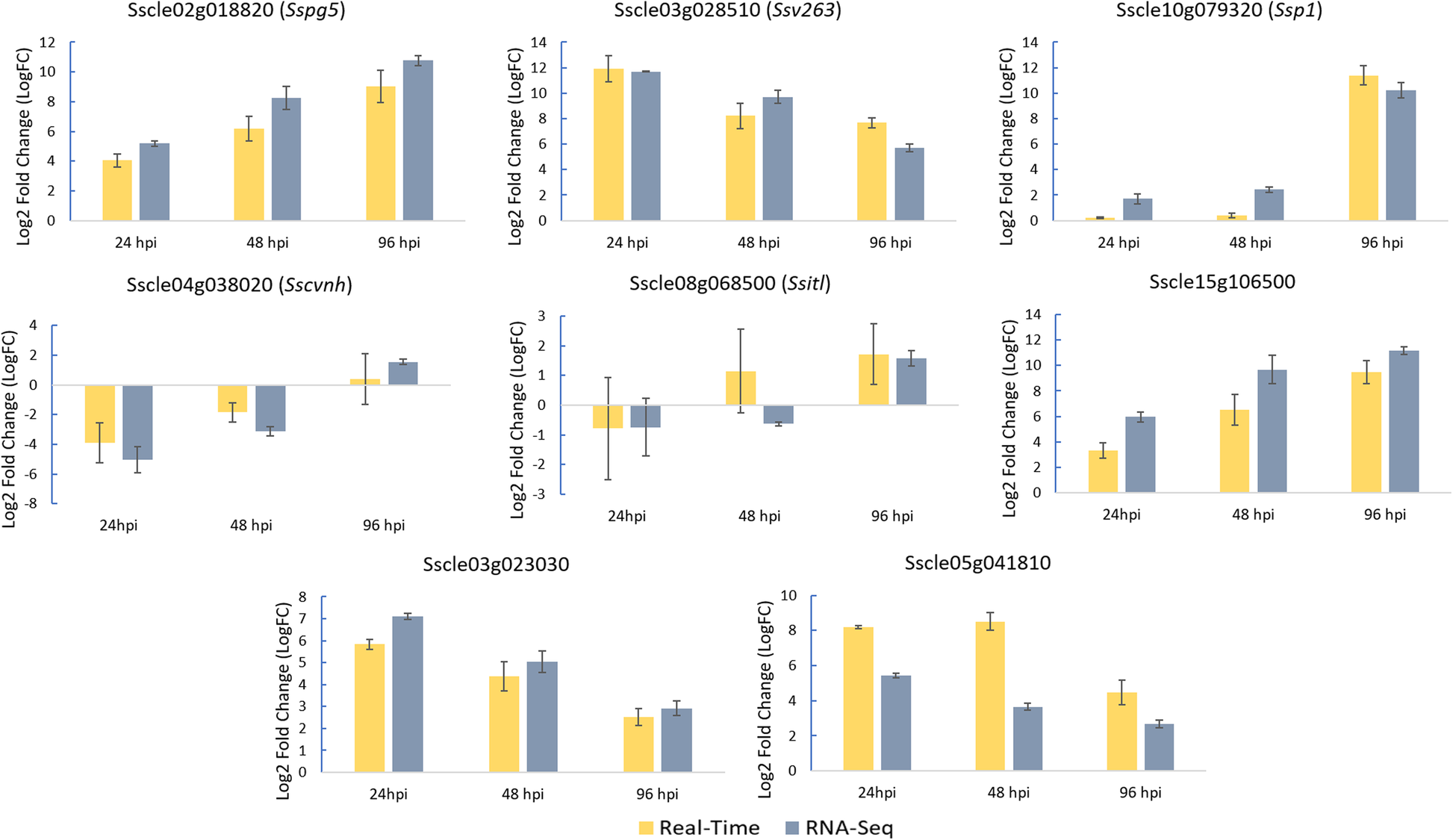
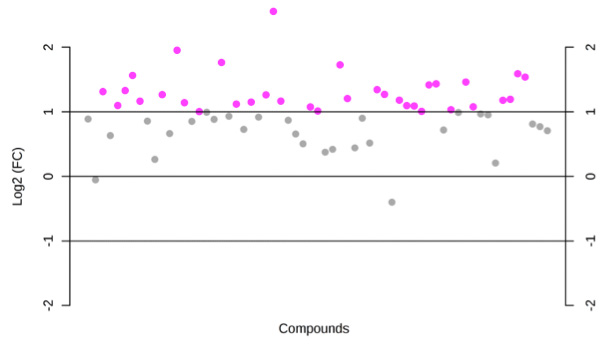
Log2-fold change values determined by qPCR for 11 markers identified as... | Download Scientific Diagram

JCI - Neutrophil gelatinase–associated lipocalin mediates 13-cis retinoic acid–induced apoptosis of human sebaceous gland cells
PLOS ONE: Genome-Wide Binding and Transcriptome Analysis of Human Farnesoid X Receptor in Primary Human Hepatocytes

Comparison of transcript expression in terms of fold change as measured... | Download Scientific Diagram

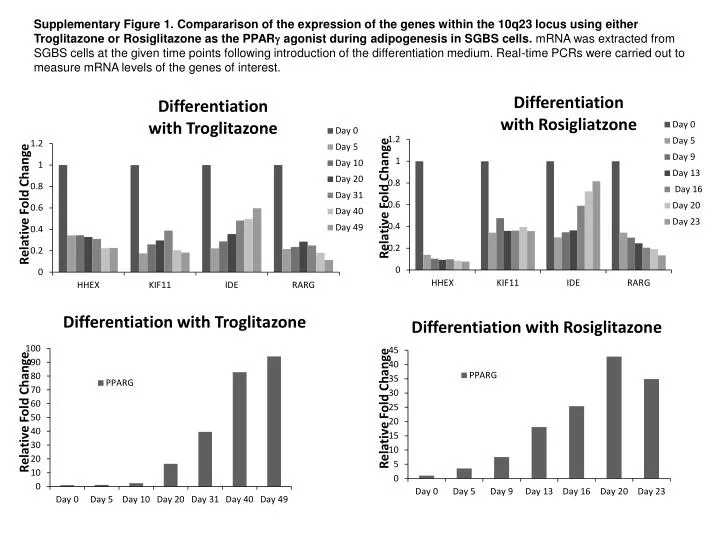
Figure S2 Fold change in gene expressions for MCF-7 cells. | Gene expression, Cell line, Cancer cell
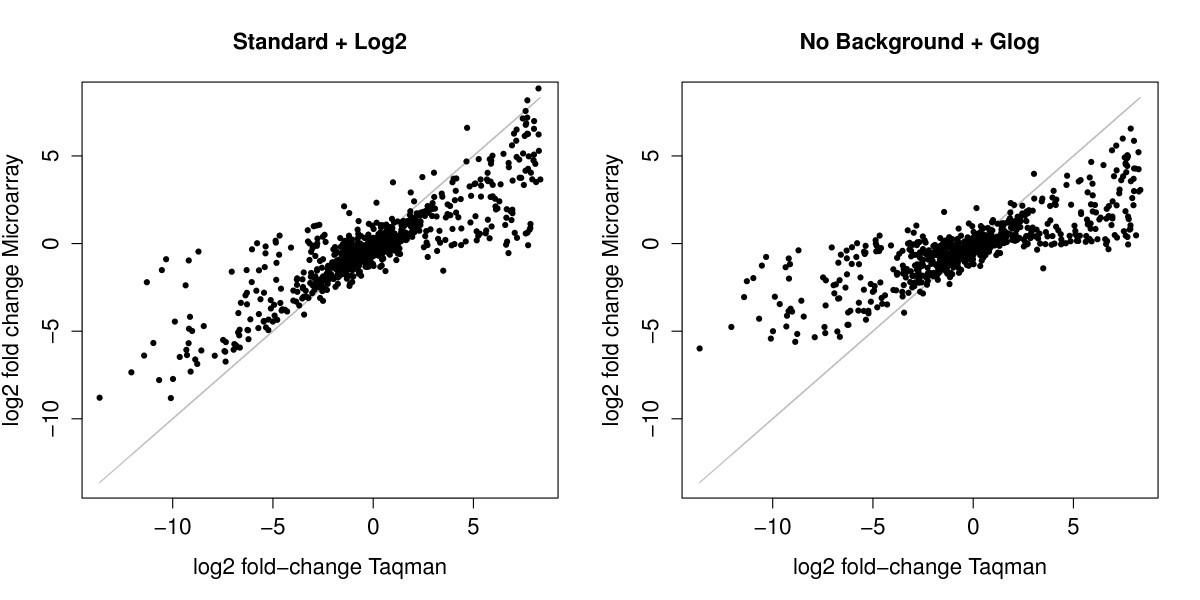
Impact of the spotted microarray preprocessing method on fold-change compression and variance stability | BMC Bioinformatics | Full Text
PLOS ONE: The NOD-Like Receptor Signalling Pathway in Helicobacter pylori Infection and Related Gastric Cancer: A Case-Control Study and Gene Expression Analyses

Graph showing relative fold change expression of inflammatory cytokines for WT and TLR4−/− mice (mean fold change over day 0; ND, not detectable).
PLOS ONE: Expression Profiling of Major Histocompatibility and Natural Killer Complex Genes Reveals Candidates for Controlling Risk of Graft versus Host Disease
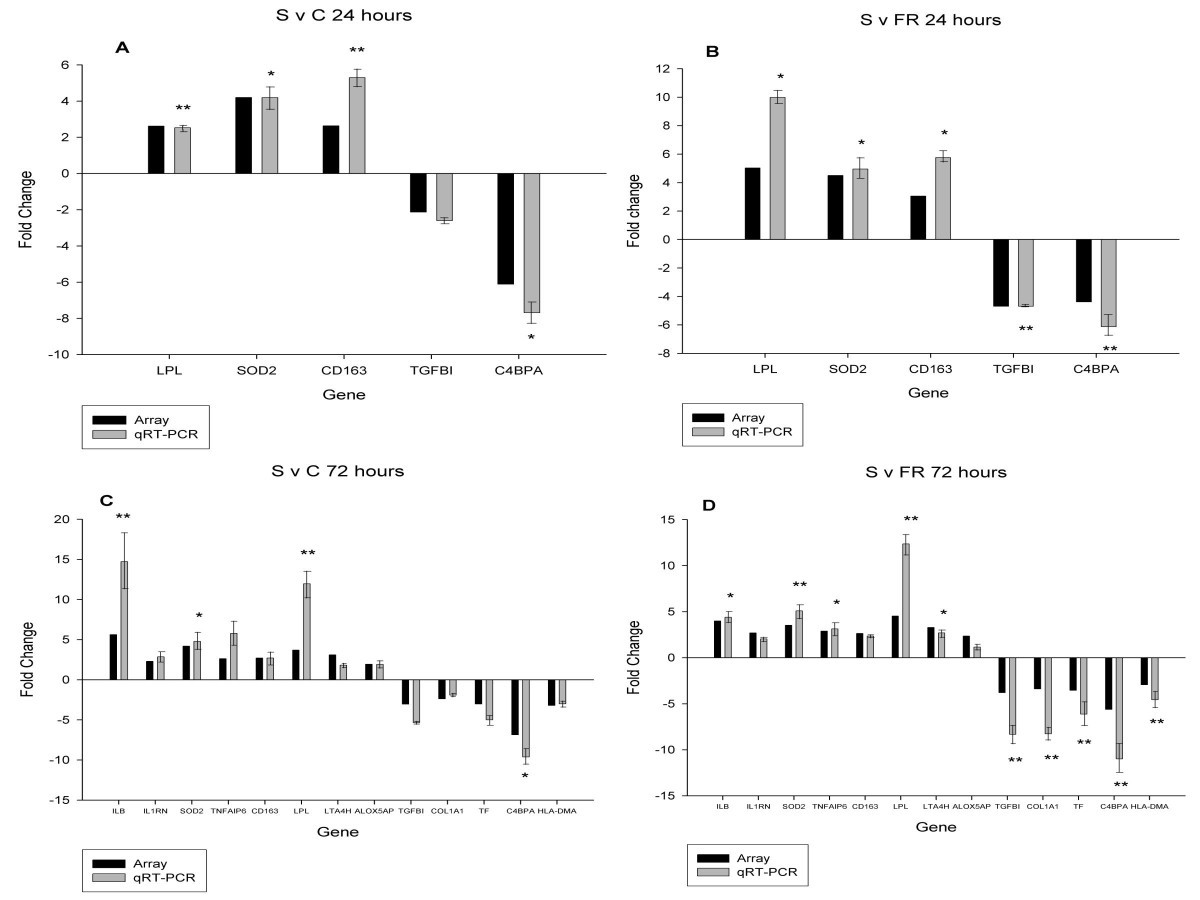
Gene expression profiling in the lungs of pigs with different susceptibilities to Glässer's disease | BMC Genomics | Full Text
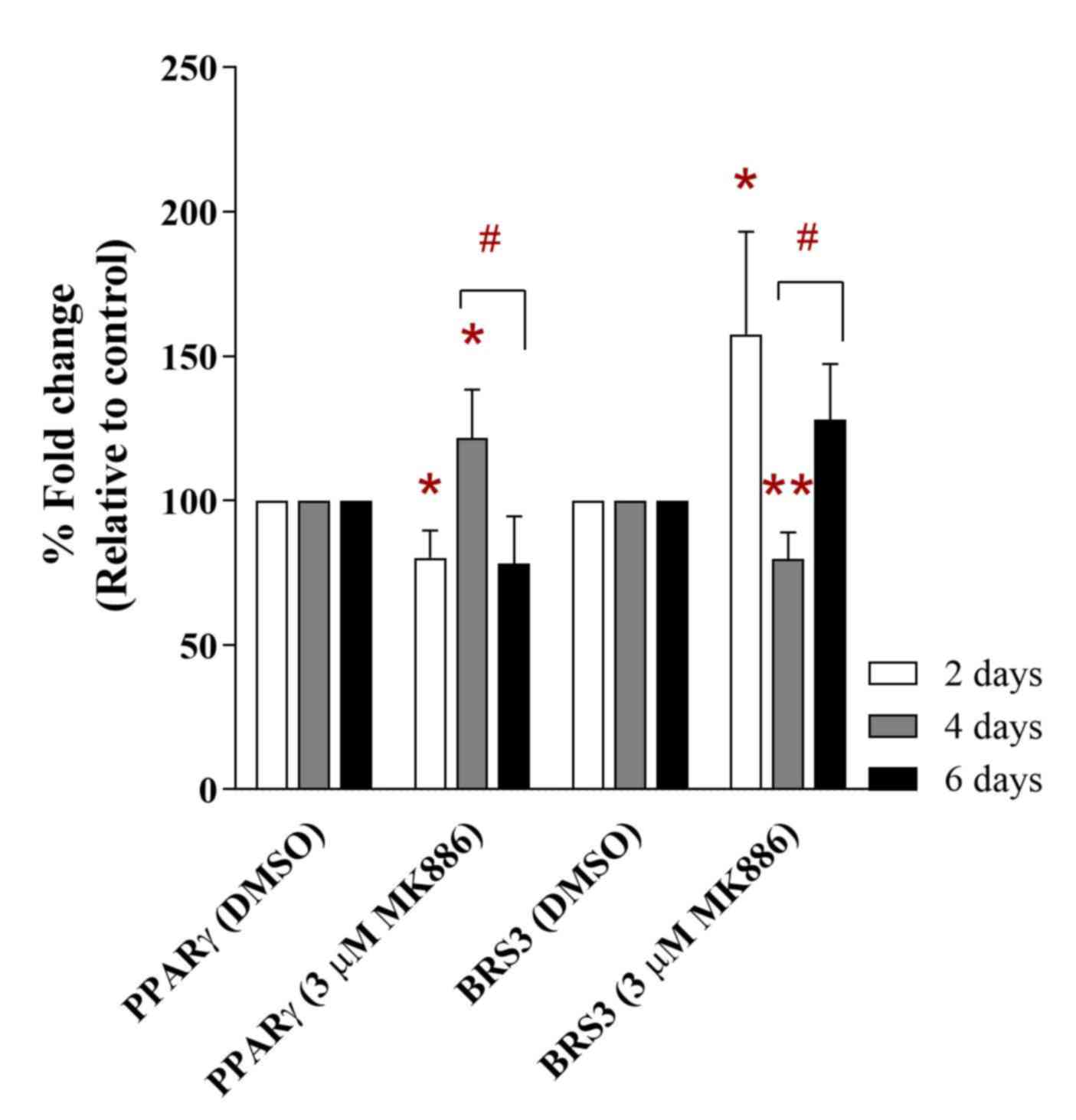
Pilot study and bioinformatics analysis of differentially expressed genes in adipose tissues of rats with excess dietary intake

Graph showing relative fold change expression of RANK, RANKL and OPG for WT and TLR4−/− mice (mean fold change over day 0; ND, not detectable).